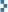This week in the online journal club #microtwjc we will be discussing this paper titled “Schmallenberg Virus Pathogenesis, Tropis and Interaction with the Innate Immune System of the Host” (Varela M, Schnettler E, Caporale M, Murgia C, Barry G, et al. (2013) Schmallenberg Virus Pathogenesis, Tropism and Interaction with the Innate Immune System of the Host. PLoS Pathog 9(1):e1003133. doi:10.1371/journal.ppat.1003133).
I’ve written before about Schmallenberg here and here so I will only go into the background briefly before diving in to the paper.
Background
Schmallenberg virus was first discovered in November 2011 so it is still pretty new to scientists. The virus infects ruminants and whilst it only causes mild disease in adult animals, if an animal is pregnant when she is infected then the baby can end up having both muscle and brain problems and can often be born dead. The virus now seems to have spread across much of Europe (see the map from FluTrackers.com). The virus itself is in the Bunyaviridae family which means that the virus is surrounded by an envelope and has a genome made up of a single strand of RNA (for reference, our genome is made up of a double strand of DNA). The virus’ genome is made of of 3 sections named (very imaginatively) small, (S), medium, (M), and large, (L). The genome codes for 4 proteins that are important in making up the structure of the virus. Schmallenberg virus is in a smaller subsection of the Bunyaviridae family called the Orthobunyavirus. Orthobunyavirus members not only have the 4 structural proteins but they also have 2 extra proteins (the NS proteins). This information will be useful for later…
So on to the paper…
Virus Growth
The first thing the authors wanted to do was to find out how Schmallenberg virus grows in various different cells – this is important because if researchers want to carry out experiments on the viruses they need to know what cells to use. The researchers tested sheep, cow, human, dog and hamster cells lines from organs including the brain, the aorta and the kidney (all of these are cell lines that were grown in the lab – they are not being tested on live animals). The virus grew in nearly all the cell lines but particularly well in sheep choroid plexus (part of the brain) cells so the researchers used this type of cell for all of the other experiments.
‘Rescuing’ the Virus
The next thing the researchers did was design a system to make more copies of the virus. (Very) basically they took the S, M and L sequences of the virus genome and put the complementary sequence into plasmids (circular loops of DNA that are found ‘naturally’ in bacteria). (So if the virus sequence was CCG the plasmid sequence would be GGC.) They then put these plasmids into another cell line and these cells replicated the plasmid genetic sequence, so making more copies of the virus.
Looking for a good animal model
Another thing the authors did was investigate whether mice could be used as an experimental model for Schmallenber virus infection. They inoculated the virus (or a negative control) into the brains of 2 day old mice. Those mice that were infected with the virus died within 8 days whereas those that had the negative control survived. They then looked at other ages, using 10 day old and 18 day old mice. Infection with the virus also killed most of the animals at these ages. When the scientists went on to look at the brains of these infected mice they found multiple abnormalities and evidence that the virus was in many of the cells.
The authors also looked at the brains of lambs and calves that had contracted the virus ‘out in the wild’ as it were. They found the virus in places very similar to where they had seen evidence of the virus in the mice, suggesting that the mice might be a good model.
Which bit of the virus makes it harmful
 A good way to find out which proteins make a disease agent harmful is to stop the virus/bacteria from producing the protein you suspect. You then infect this mutant back into your cell lines or animal model and look to see if disease still occurs. If the disease is no longer seen then you know that protein is vital for the disease (although it is possibly not the only one involved). If the disease proceeds exactly as it did before you know that you were wrong in your suspicion.
A good way to find out which proteins make a disease agent harmful is to stop the virus/bacteria from producing the protein you suspect. You then infect this mutant back into your cell lines or animal model and look to see if disease still occurs. If the disease is no longer seen then you know that protein is vital for the disease (although it is possibly not the only one involved). If the disease proceeds exactly as it did before you know that you were wrong in your suspicion.
The scientists who wrote this paper did just that – they ‘knocked out’ one of the non-structural (NS) proteins and infected mice with the strain. All the mice inoculated with the original strain died within 9 days (so very similar to the previous experiment), all the control animals survived. Of the mice infected with the mutant strain, deaths that occurred happened later than they did in the original strain group and 40-60% survived until the end of the experiment. These results suggest that the NS protein has an important role to play in the disease caused by the virus, although it is obviously not the only one involved.
To investigate this further the scientists first went back to previously published scientific literature – the NS proteins of other viruses in the same group have been shown to inhibit the making of host immune system proteins called interferon (IFN) alpha and beta. The scientists speculated that this might also be true for the NS protein that they knocked out. To test this they first infected some cells that can produce IFN with the Schmallenberg virus (either the mutant or the original strain). They then collected the supernatant (basically the fluid on top of the cells). If their hypothesis was correct then the fluid from the original strain-infected cells would contain not contain any IFN (because the NS protein inhibits its production) but the fluid from the mutant-infected cells would. They tested the fluid in a very clever way. They got another cell line, this time one that could not make IFN. They added the fluid from the first set of cells. Then they infected these new cells with a 2nd virus. This 2nd virus is susceptible to IFN so if the supernatant contained IFN the virus would not be able to properly infect the cells whereas if the supernatant did not contain IFN then the viruses would infect the new cell line.
The supernatant from the original strain-infected cells must not have had IFN because the 2nd set of cells got infected with the 2nd virus. In contrast the supernatant from the mutant-infected cells must have had IFN because the cells did NOT get infected with the 2nd virus. And so this tied in nicely with the scientists’ prediction.
Finally, to confirm this the scientists infected the 2 virus strains into mice that could not respond properly to IFN. This time 80% of the original strain-infected mice died within 4 days and the rest died within the next 2 days. ALL of the mutant-infected mice died within 4 days. This further suggests that an IFN response is vital for protection against disease caused by Schmallenberg virus.
And that’s where this paper ends…
… having covered a lot. We now know what type of cells we can grow the virus in, we have a proposed animal model of the disease and we know a little (tiny) bit about what the virus needs to cause disease and what can protect the host. The paper also has the details of 2 different ways a synthetic virus can be produced (see the ‘rescuing’ section) which sounds like it will be of great help to researchers in the future.
I thought it was a very interesting paper and I am looking forward to discussing it with anyone and everyone who joins us at journal club on Tuesday 22nd January 8pm GMT (just follow the hashtag #microtwjc on Twitter).
Images
The map is kindly borrowed from FluTrackers.com
The ‘When germs go bad’ (one of my favourite microbe pictures) is by Gaspirtz (Own work) (made available under creative commons licence)
The lamb is by Evelyn Simak (made available under creative commons licence)
References
1.Varela, M., Schnettler, E., Caporale, M., Murgia, C., Barry, G., McFarlane, M., McGregor, E., Piras, I., Shaw, A., Lamm, C., Janowicz, A., Beer, M., Glass, M., Herder, V., Hahn, K., Baumgärtner, W., Kohl, A., & Palmarini, M. (2013). Schmallenberg Virus Pathogenesis, Tropism and Interaction with the Innate Immune System of the Host PLoS Pathogens, 9 (1) DOI: 10.1371/journal.ppat.1003133
![]()